Regulation of lignin biosynthesis: Key factor for lignocellulose-based value added chains
The fast growing C4 grass Miscanthus has a high biomass yield potential even on marginal soils and is therefore highly suitable as a future biomass crop. Biomass composition is of fundamental importance for various applications. Main components of plant cell walls include the polymers cellulose, hemicellulose and lignin. The lignin component is an important research objective as it needs to be separated from the sugar polymers to make the latter available for biochemical and chemical conversion. However, lignin could also serve as a resource for aromatic platform chemicals, if ongoing studies to produce functionalized lignin with well-defined characteristics will be successful. Thus, cultivating Miscanthus with increased or lowered lignin content is of great interest. Consequently, understanding the regulation of lignin biosynthesis in Miscanthus is of high priority.
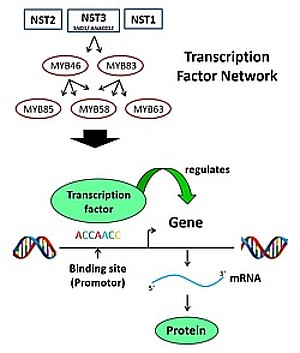
The aim of this subproject was to investigate the transcriptional regulation of lignin biosynthesis in Miscanthus sinensis. Lignin biosynthesis is regulated by a complex network of transcription factors (TF), the expression of which is modulated by various cultivation parameters. Using sequence information from Arabidopsis, maize and millet, candidate genes coding for TF in M. sinensis were identified using an RNA-seq-database. Candidates for all three lignin biosynthesis regulating levels were included (level 3: SND1, master regulator for secondary cell wall formation, level 2: MYB46/MYB83, level 1: MYB85/MYB58/MYB63 - gene denomination in accordance with Arabidopsis nomenclature), but also two repressing TF (MsMYB31 and MsMYB42). The cell wall regulator MsSND1 was further studied with respect of its functionality. The Arabidopsis double mutant nst1/nst3 could be complemented with the MsSND1 gene, and, using a DEX inducible MsSND1 gene construct, direct and indirect target genes could be identified.
Partial sequencing of the M. sinensis genome provided a database for cloning the promoter sequences for both the transcription factors and the lignin biosynthesis genes. Transient expression of TF genes in tobacco leaves (or in grapevine cells, using the dual luciferase assay) showed that all candidate TFs activated the promotors of the predicted target genes of lignin-biosynthesis, among which the promoter of the laccase gene MsLAC1, coding for an enzyme which upon overexpression can modify lignin content and composition. In collaboration with the research group of Shawn Mansfield (UBC, Vancouver), the influence of these transcription factors on the qualitative lignin composition (S-/G-lignin, etc.) was demonstrated.
| Project title | Regulation of lignin biosynthesis: Key factor for lignocellulose-based value added chains |
| Institution | |
| Research group | Prof. Thomas Rausch, Dr. Sebastian Wolf, Philippe Golfier, Feng He, Wan Zhang |
| Project status | completed |
